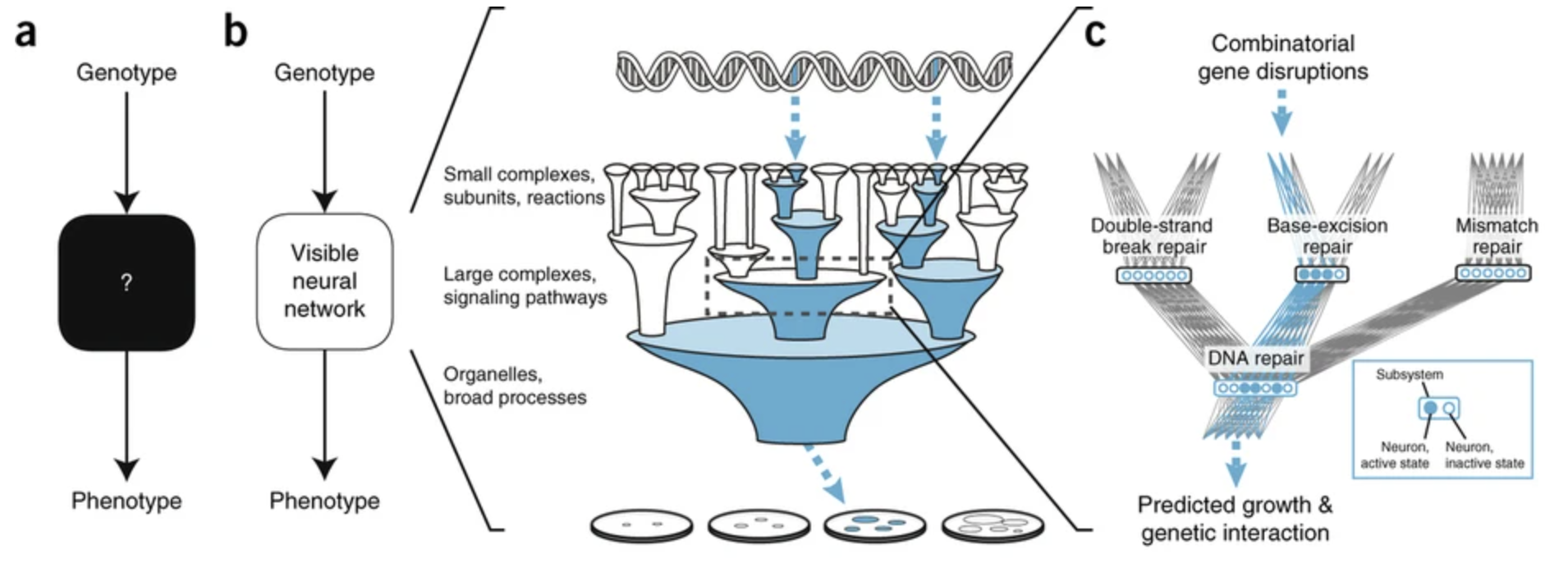
In DCell, the functional state of each subsystem is represented by a bank of neurons (Fig. 1c). Connectivity of these neurons is set to mirror the biological hierarchy, so that they take inputs only from neurons of child subsystems and send outputs only to neurons of parent subsystems, with weights determined during training. The use of multiple neurons (ranging from 20 to 1,075 per system; see Online Methods) acknowledges that cellular components are often multifunctional, with states that are too complex to be captured by a single neuron. The input layer of the hierarchy comprises the genes, whereas the output layer, or root, is a single neuron representing cell phenotype.
DCell trains a deep neural network to predict phenotype from genotype, with architecture that exactly mirrors the hierarchical structure of an ontology of cellular subsystems. Each cellular subsystem is represented by a group of hidden variables (neurons) in the neural network, and each parent–child relation is represented by a set of edges that fully connect these groups of hidden variables.
References: Using deep learning to model the hierarchical structure and function of a cell